noise-free-cnv
noise-free-cnv is a program for analyzing and manipulating DNA microarray data. It is written in C++, using Gtkmm and FFTW.
noise-free-cnv is free software under the terms of the GNU General Public License version 3.
Downloads:
Motivation
The noise-free-cnv program package has been specifically developed to analyze DNA microarray data for copy number variation and to manipulate such datasets in order to reduce noise.
The central program of the noise-free-cnv package is noise-free-cnv-gtk, a visual editor for interactive visualization and manipulation of DNA microarray data. Besides functioning as a browser for direct inspection and verification of alleged CNV findings, it allows the user to perform many operations on the data to separate and eliminate noise components.
As a second program, noiser-free-cnv-filter implements a specific algorithm for system noise reduction of DNA microarray data. It is usable as a command line program to be easily applied to a batch of datasets.
Both programs are able to read and write files suitable for PennCNV.
Features
- interactive visualization of DNA microarray data
- provides functions to enhance the quality of the data
- compatible with PennCNV files
- easy to install and to use, no special hardware or software requirements
- available for Windows (XP, Vista, 7) and Linux (Debian, Ubuntu, Mint)
Examples
Subtracting files from each other
| 1) start the program | 2) load a file containing LRR and BAF values | 3) visualize the file |
 |
 |
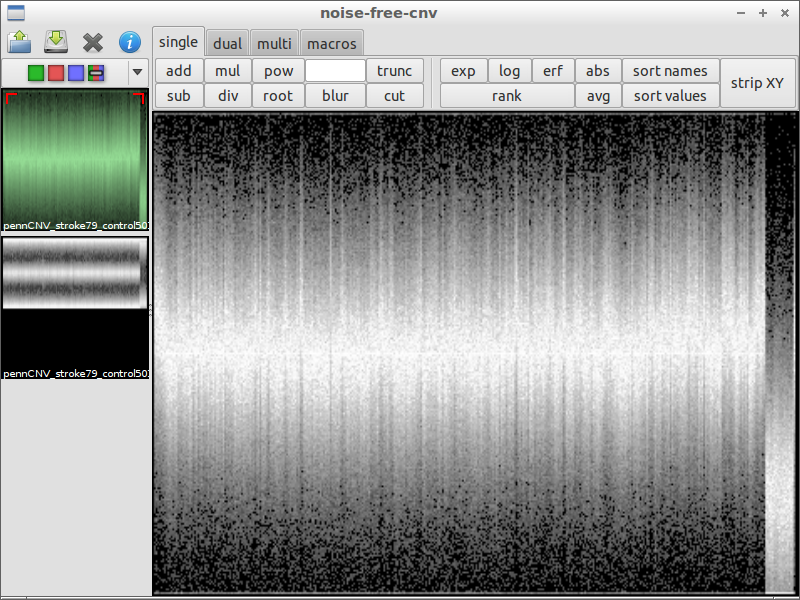 |
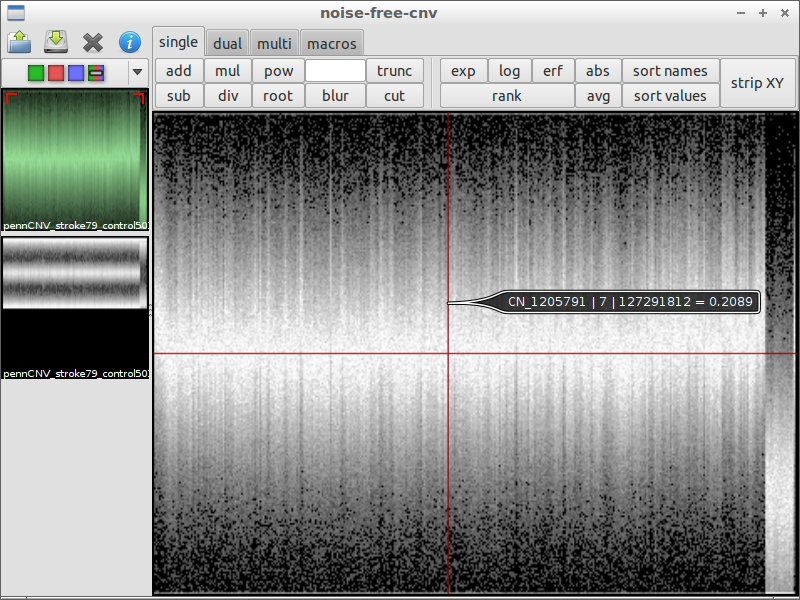
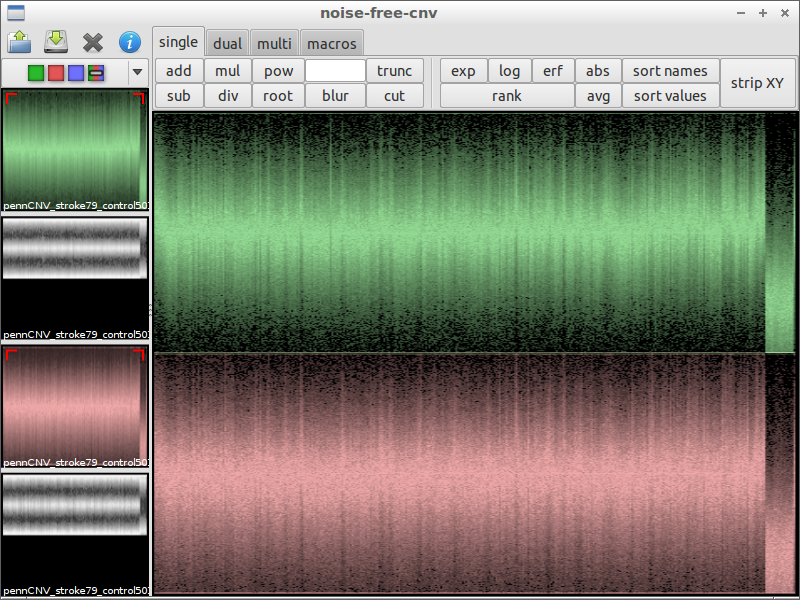
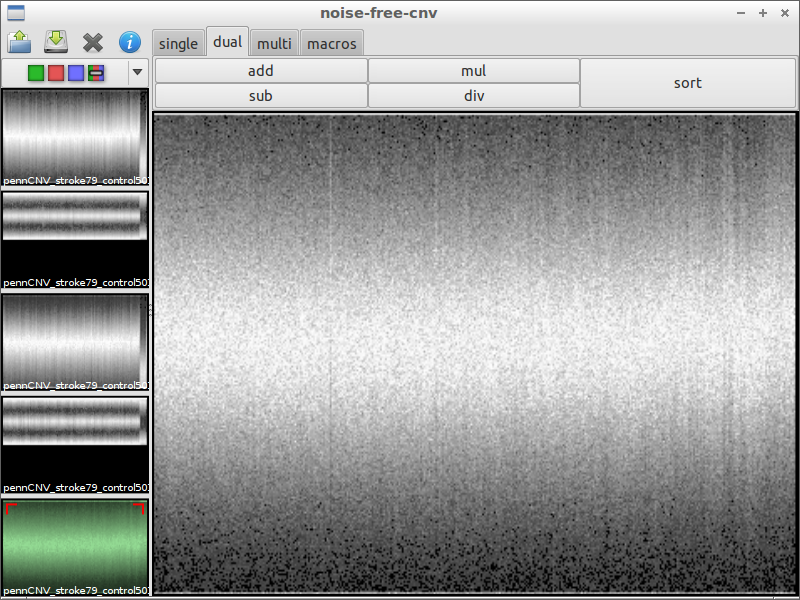
| 4) inspect interesting spots | 5) load another file for comparison | 6) subtract the files from each other |
 |
 |
 |
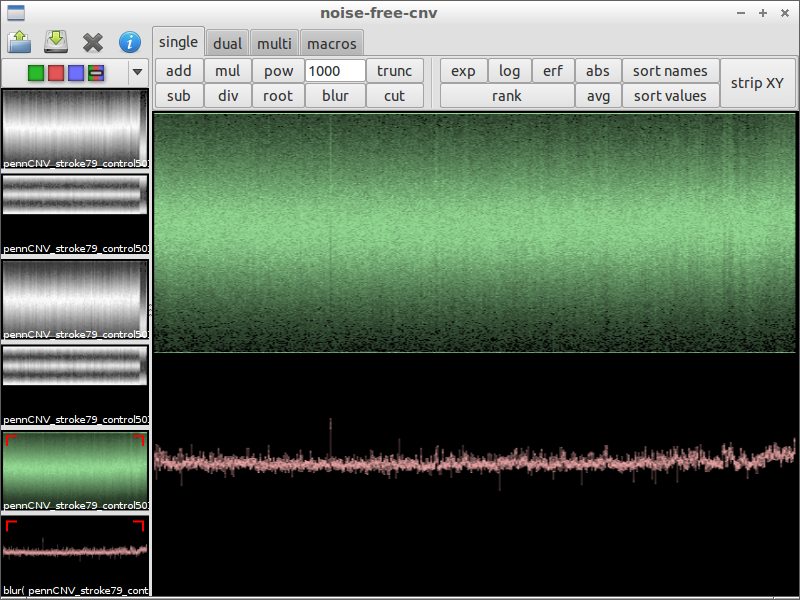
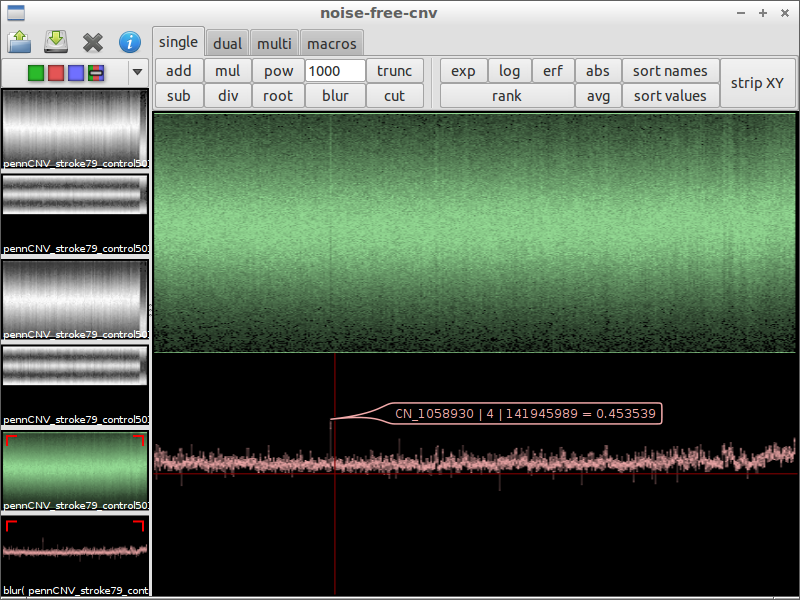
| 7) use blur to remove noise | 8) look for irregularities | 9) zoom in and compare with the original data |
 |
 |
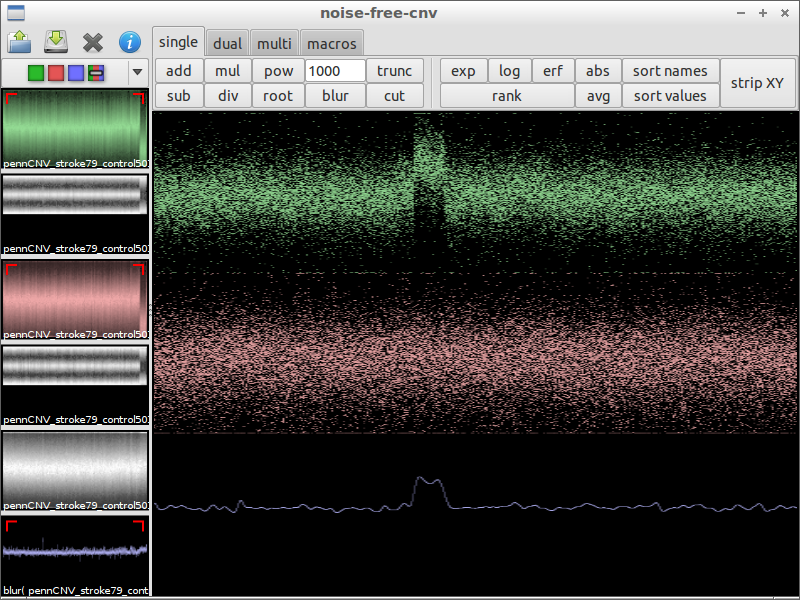 |
Filtering high and low frequencies
| 1) start the program | 2) load a file containing LRR and BAF values | 3) use blur with a huge factor to get the chromosomal waves |
 |
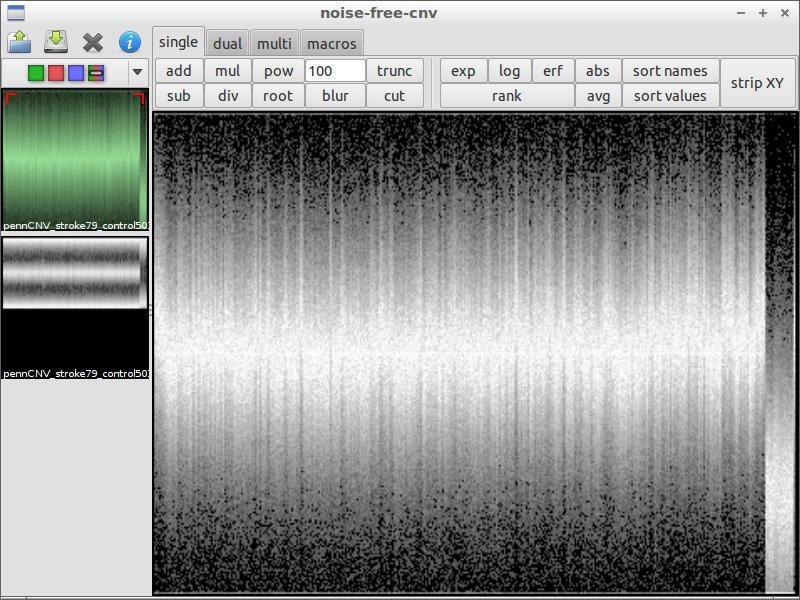 |
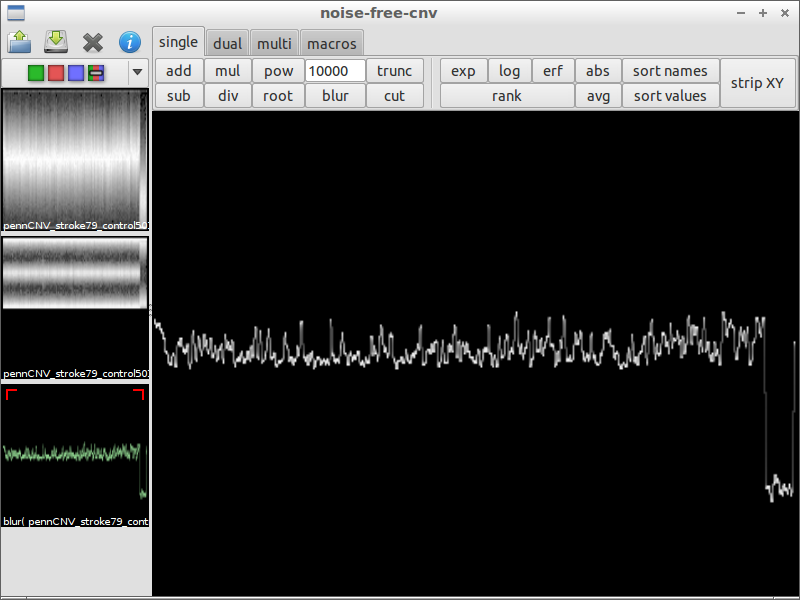 |
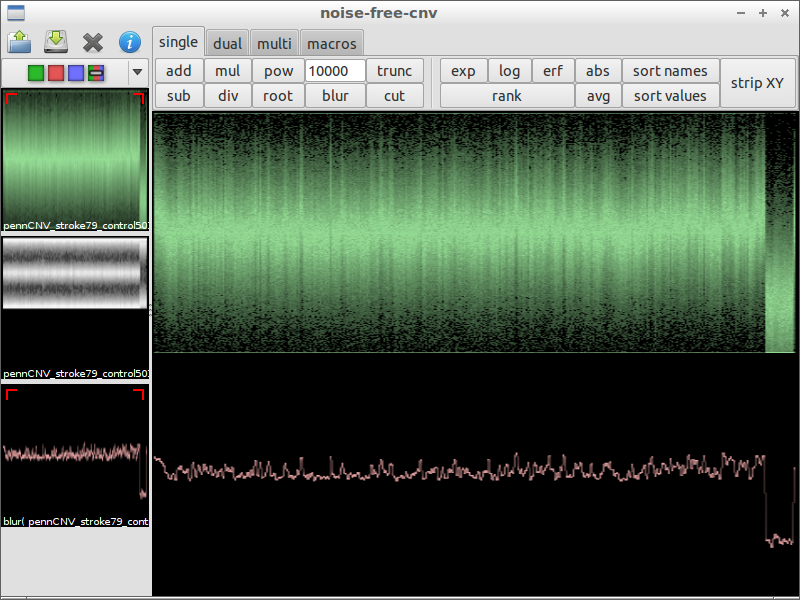
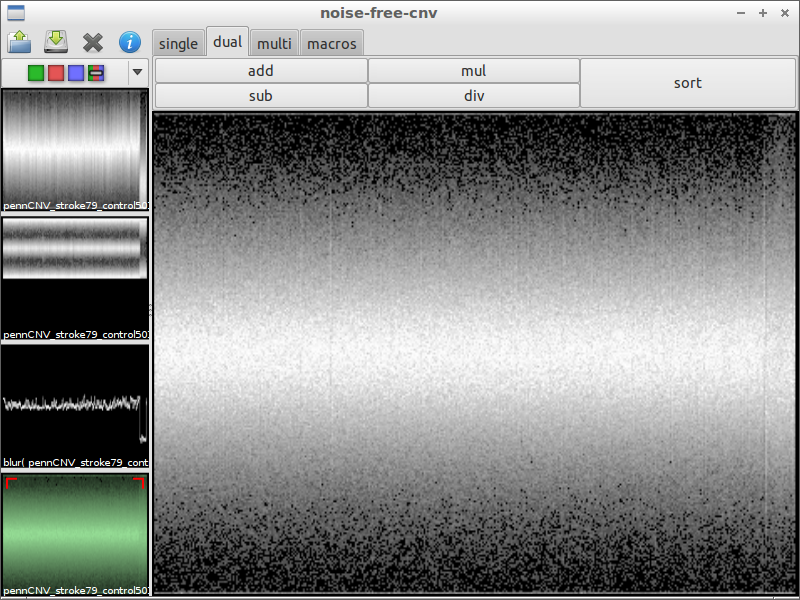
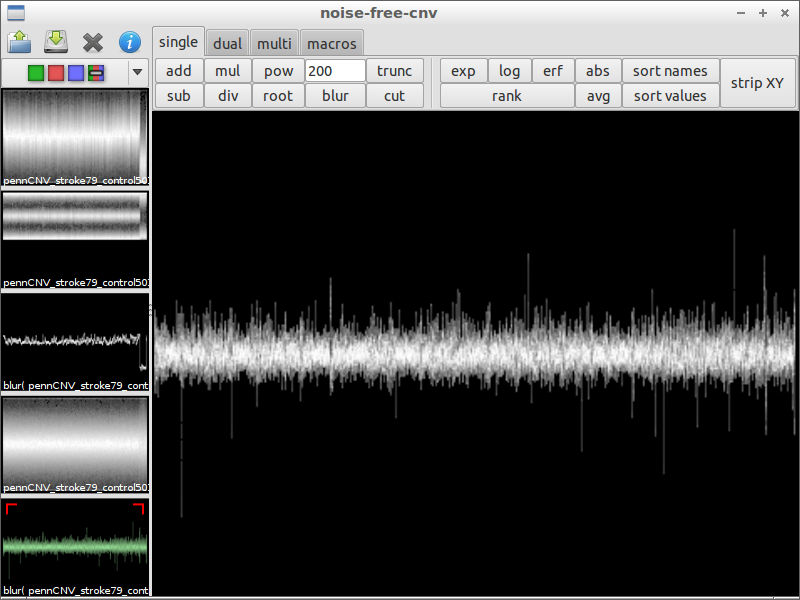
| 4) compare with original data | 5) subtract the chromosomal waves | 6) use blur with a small factor to remove noise |
 |
 |
 |
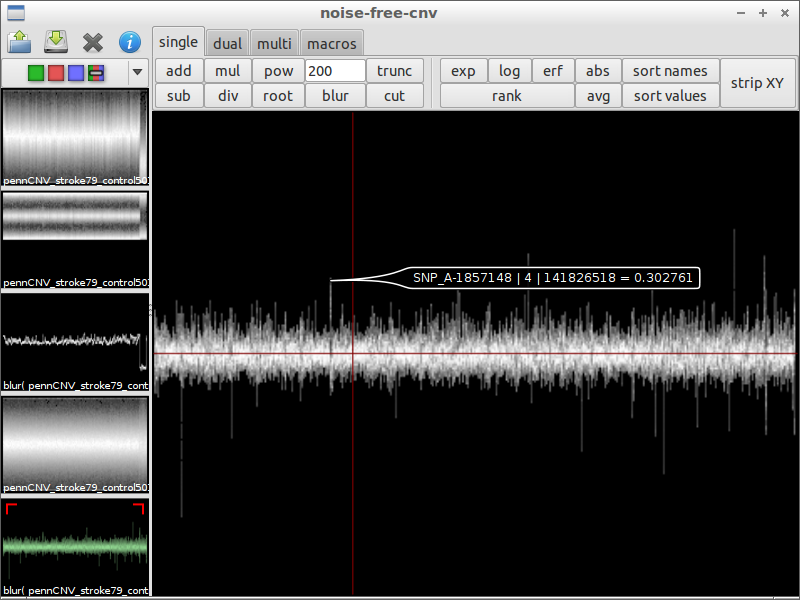
| 7) search interesting spot | 8) compare with the original data | 9) zoom in |
 |
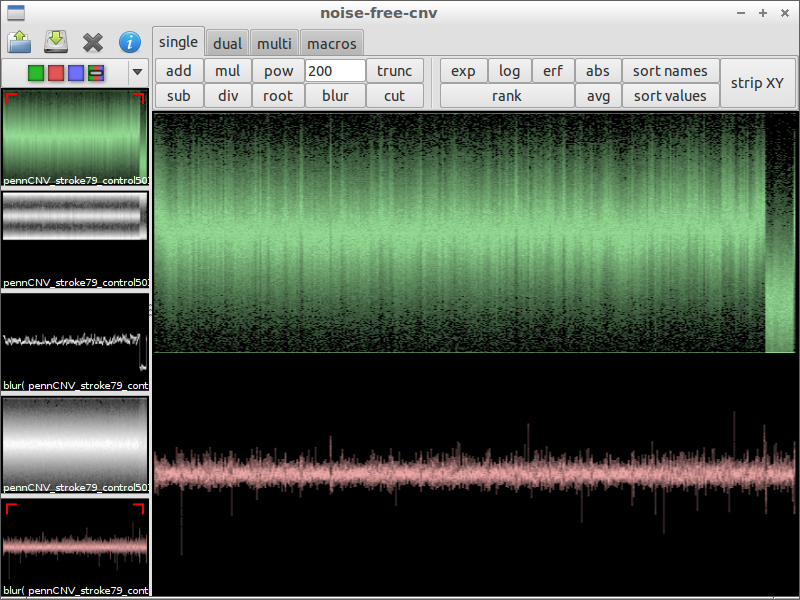 |
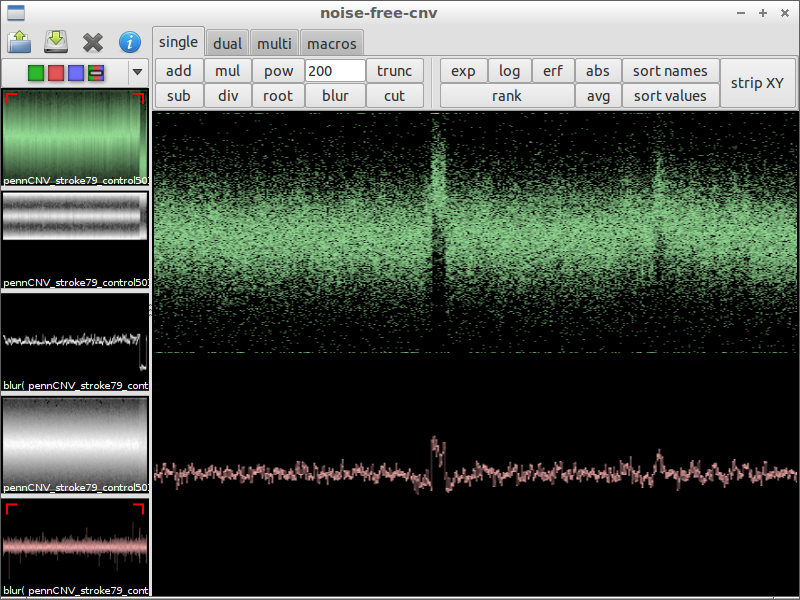 |